As my first research project as a student at Abilene Christian University, I was a member of the AP5 research team. The goal of our research was to confirm and further develop a newly constructed phylogeny of rodents in the genus Thomasomys. A phylogeny is a hypothesis of the evolutionary history and the evolutionary relationships between species. The reason that it is important to test the previously constructed phylogenetic tree is because a phylogeny is always a hypothesis. The more DNA that is tested, the more confident we can be of the likelihood of our hypothesis. A fully resolved tree (where the exact relationships of all of the species are described) is important because it can be used to understand the impact of historical events like the formation of the Andes on the diversification history (how species separated) of Thomasomys. The phylogeny that has already been constructed was found using mitochondrial DNA, and our research was attempting to test the mitochondrial phylogeny with nuclear DNA. The mitochondrial genome comes from organelles called mitochondria (inherited from the mother) and contains anywhere from 100 to 1000 copies per cell while nuclear DNA is found in the nucleus (inherited from both parents) and contains two copies per cell.

Thomasomys caudivarius
In order to collect DNA from the AP5 (intron 2 from the acid phosphatase type V gene) region, we conducted polymerase chain reaction (PCR) on DNA samples from species of Thomasomys with primers that we learned about from previous research on rodent phylogenies. PCR is used to amplify a piece of DNA until there are millions of copies of the DNA sequence. DNA polymerase is the enzyme that replicates the DNA, and primers are short strands of DNA that are used to facilitate DNA replication because the polymerase requires an existing strand of DNA to which it can bind the replicated DNA. However, these primers didn’t work very well with Thomasomys because they were designed for other rodent species, but we were able to amplify AP5 from a few of our samples. We used the sequence that we obtained to design three new sets of primers, and two of the three sets were successful at amplifying AP5 from most of our samples. After the DNA is amplified using PCR, the product of the PCR is then tested to see whether our targeted area was present with gel electrophoresis. Electrophoresis separates different sizes of DNA strands. If there is DNA of the correct size present after the gel, then we assume that the AP5 region is probably present, and this DNA will then be cleaned and sent off to be sequenced. This sequence is what is then used to estimate evolutionary relationships between species by looking at how the sequences vary from each other.
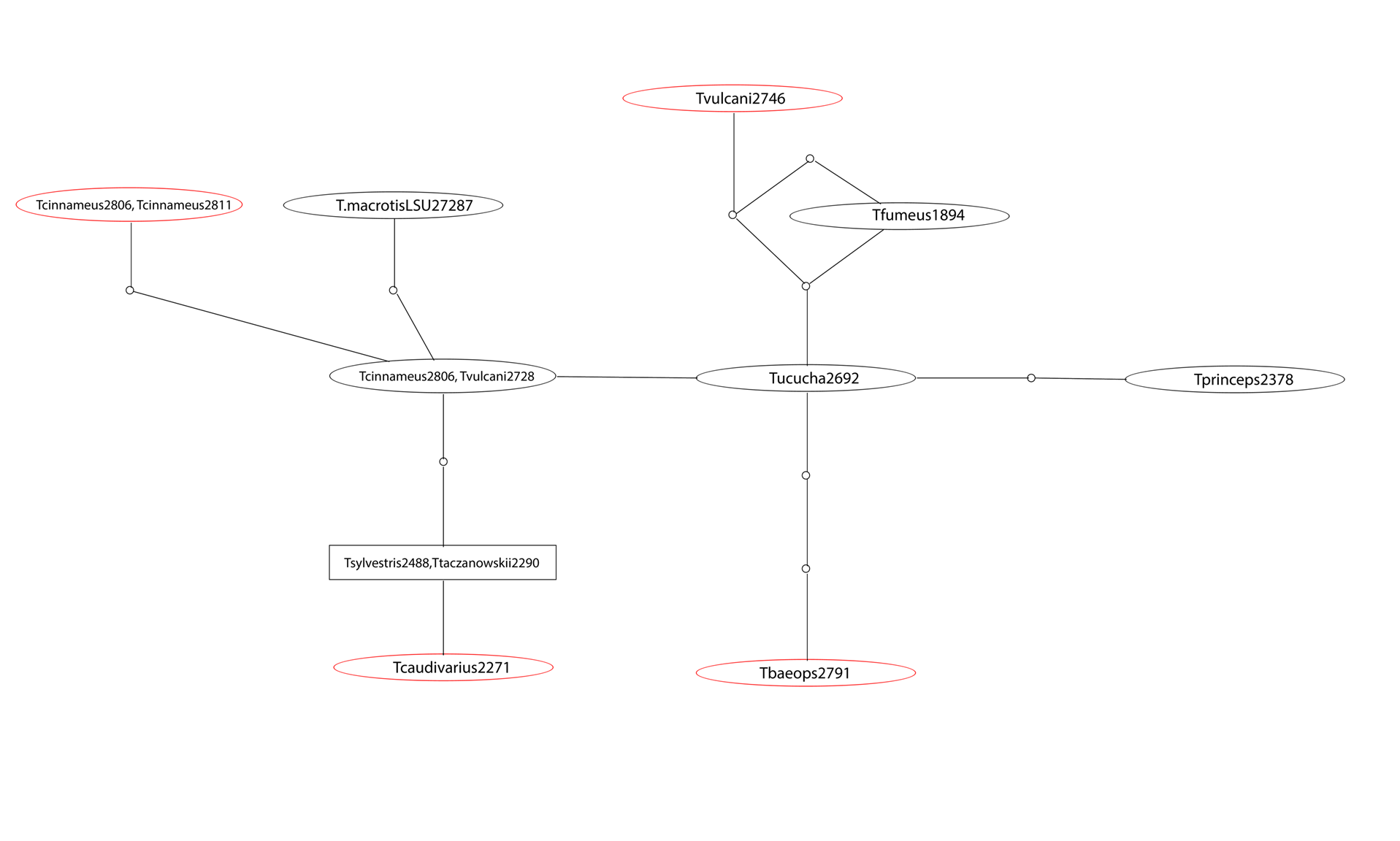
Figure 1: Genotype network representative of the AP5 sequences collected. The segments in between each oval represent a different mutation. The black ovals represent the results from primers used initially, and the red ovals represent the results from primers designed in our own labs. Although the red samples are related in a pattern congruent with mitochondrial hypotheses, they differ by only a small number of mutations in AP5.
Sequences from the AP5 gene have been both easier to obtain and easier to interpret and edit after the change in primers. The resulting sequences from our new primers resulted in a preliminary phylogeny that was compatible with phylogenies based on mitochondrial DNA. However, the AP5 gene did not help create a more resolved phylogeny due to the lack of variability in the AP5 marker compared to the mitochondrial markers and other nuclear markers like RAG1. Since our project began, we have learned of more promising nuclear markers that we will focus on in the future.
Other than the lack of variability of the AP5 gene, there are other potential causes of the lack of success with AP5 such as: human error in running the PCR or gel electrophoresis, the primers, or the temperature of the PCR amplification. As my time as a researcher on the AP5 team progressed, the amount of successful AP5 PCRs detected by electrophoresis increased gradually. This lends me to believe that human error caused by inexperience may have played a small role in difficulty of obtaining results. My skills in conducting PCR and gel electrophoresis improved as a result of this research, but more importantly I gained experience in applying knowledge learned in class to the real world. As a recent transfer to ACU, being a part of ACU biology research was an incredible way to feel included at a new university. There is a sense of community within research at ACU where students help each other learn and grow in a comfortable environment, which is a unique aspect not found at every institution. Towards the end of the semester, we participated in the ACU Undergraduate Research Festival with a poster presentation. As part of this presentation ACU professors circled the room asking questions of the presenters in order to test their knowledge of the research. This was my first experience at a research conference, and, being new to research, I was extremely fearful. Although initially this was a difficult experience for me, by the end I was grateful that I was a part of it. Hearing the more experienced members of our team answer questions gave me a far better understanding of our research, and I will be able to use this experience in the future.

