Editor’s Note: This blog was composed through a research collaboration by Marissa Horne and Cameron Ludwig.
Marissa Horne:
The Abilene Christian University Biology Department caters to a host of professions. We, the students learning from these amazing professors, have been given the incredible opportunity to assist in research projects by working and learning under the guidance of our teachers. This was the case with me. One semester ago, I approached Dr. Joshua Brokaw because I was interested in doing some entry-level research as a springboard to start working on more in depth and more complicated projects in the future. As it turns out, what I thought would be entry-level research was an ongoing project that was anything but easy. I started learning the process under some upperclassmen that had worked with the material before. It felt like a guided sink or swim lesson in DNA sequencing. I eventually learned, and by my next semester I was able to help a friend of mine learn the same methods and practices that would allow us to work together on the RAG1 (recombination activating gene 1) Thomasomys project (Thomasomys is a genus of South American rodents).

Thomasomys sp. – photo by Jorge Brito M.
Learning curve aside, the process and methods that we utilize in the lab are very interesting, and executing them well is crucial to obtaining good results. Every week that we came into lab we were either running a PCR or a gel electrophoresis. There is always much more to it than that, but as my fellow researchers can concur, this was the gist of daily lab work for this project. We definitely had some blunders; when two people with limited research experience are put together, some mistakes are bound to be made. However we were able to fix what needed to be fixed, and get decent results. I very much enjoyed seeing my own progress and confidence in the lab grow as the semester went on. My first time working on this project it was easy to let the more experienced researchers take the lead. This year I was the more experienced researcher, so I really had to be sure of what we were doing before we did it. The whole experience could pretty much be summed up in the word, learning. We are learning new things about our research subjects, we are learning how to conduct a proper research study, and we learn the valuable lesson of patience over and over again. Our project was almost a repetition, more like a confirmation, of previous hypotheses about relationships within Thomasomys. There was a lot of waiting and repeating steps, over and over again, in many aspects of this experience.
Fall 2016 Biology Research Poster Session
Towards the end of the semester, we began to prepare for our poster board presentation. Here we needed much guidance from Dr. Brokaw to put together a phylogenetic tree that showed our most recent results. This was the step that basically put together all of the small details into one big picture, or poster! One of my favorite parts of this research experience is getting to do the research festivals and the poster presentations, because I get to share a part of this interesting experience with others. I especially enjoyed talking to the first year students. They had prepared questions to ask us, but most of the time the more curious students ended up asking more than what was required of them. These events are where future researchers are found, so I really had fun talking to them about the work we had been doing.
Overall I can confidently say that Biology Research this semester was one of the most enjoyable and informative classes that I took. It was a time that I didn’t have to worry about anything else besides the task at hand. I can’t wait to see where this project will lead, and I’m hoping to continue doing research in the future.
Cameron Ludwig
I was one member of the team researching the RAG1 gene in Thomasomys. The ultimate goal of our research was to attempt to further develop the phylogeny that has been formed for the genus. A phylogeny is a hypothesis that attempts to explain the evolutionary history and certain relationships between different species sort of like a human geneology. The issue is that the phylogeny is simply a hypothesis. Our goal as researchers was to test out the hypothesis based on data that has already been produced by collecting new data to hopefully confirm previous findings. The goal of the research is to increase confidence that this phylogeny does accurately show the relationships within this genus. When we sequence more DNA, we will be able to gain more confidence in the phylogeny and more fully explain the relationships. One of the issues with the previously sequenced DNA was that previous teams have been using mitochondrial DNA which is only accurate in showing the phylogeny through the female lineage. Our research was important because we were using nuclear DNA that will help to further expand that phylogeny by including the female and male lineage.

Our main tool for collecting DNA sequence data was the PCR machine (a.k.a. thermal cycler). The thermal cycler utilizes the polymerase chain reaction in order to copy segments of DNA for analysis. Once we produced our data from the PCR, we separated and identified the different sizes of DNA strands using gel electrophoresis. Once those strands were separated, we were able to send purified DNA for sequencing and use this to form our phylogenetic trees and further understand the evolutionary relationships present.
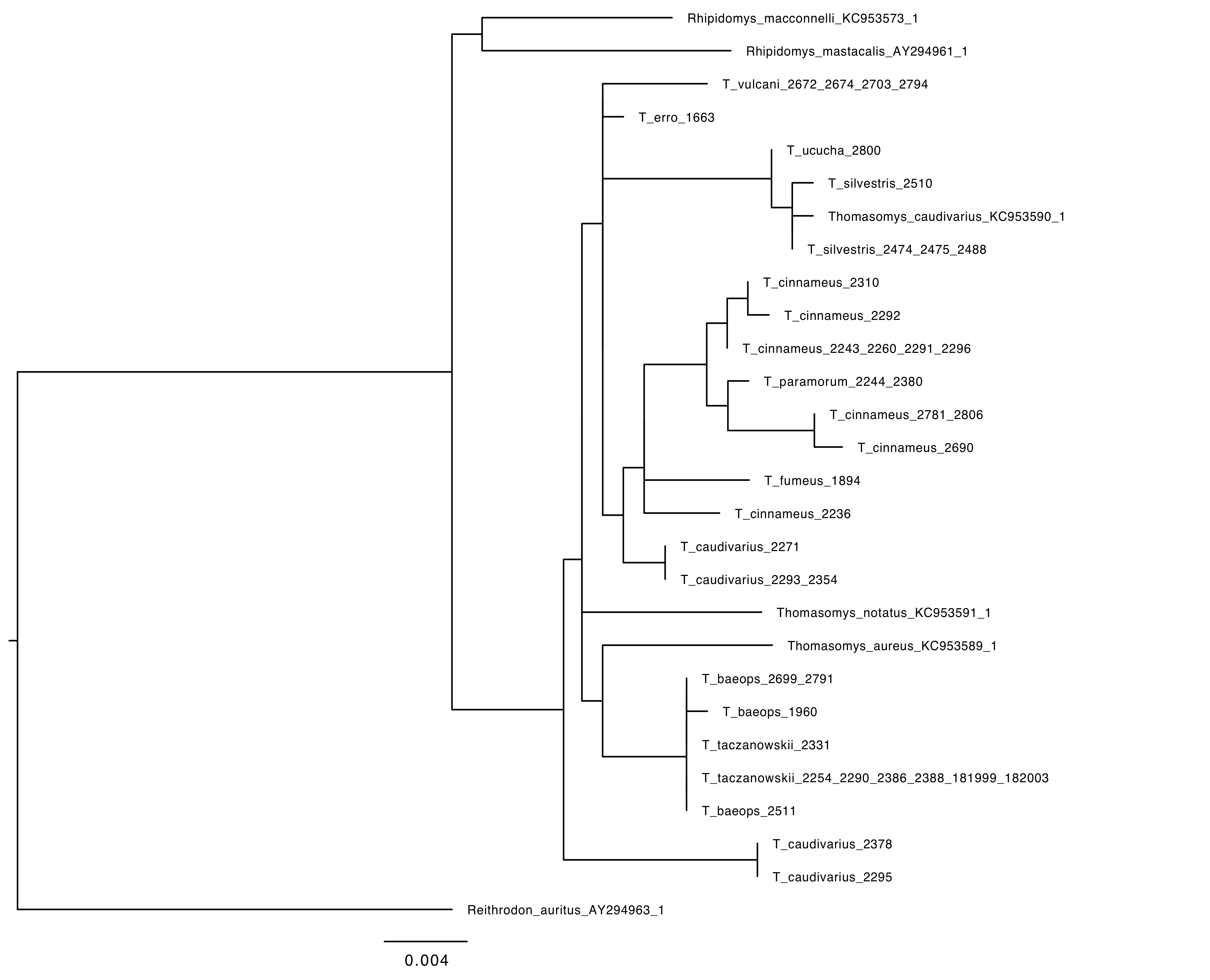
Phylogeny of Thomasomys based on RAG1
Overall, I had a wonderful experience in the lab with John, Marissa, and Dr. Brokaw. I gained more understanding of what it takes to be successful in a laboratory. I learned from my colleagues and from the mistakes that I made and will use the experiences and knowledge that I have gained this semester to be a better researcher next semester.
