This month we made a huge step toward our short term goal of sending off the Thomasomys DNA samples for sequencing. Dr. Brokaw and I found a set of primers that could amplify an approximately 1200 base pair region of the cytochrome-b gene that we are working with. We tried several primer sets and had to run a large number of PCRs in order to determine which primer set gave us the best amplification results with the largest number of our rodent DNA samples. One step in our progression towards determining the best method for amplification was running a gradient PCR to try the reaction at various temperatures (the same DNA sample is run at a number of different annealing temperatures), through this we learned that a slightly lower annealing temperature (46° C) than our previously used temperatures (50-54° C) gave us the best results.
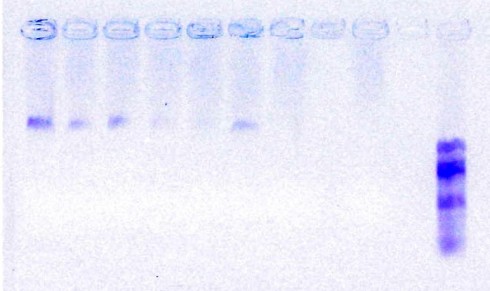
Gel of gradient PCR performed on a single DNA source amplified using the p484-p485 primer pair. Annealing temperatures from left to right were 44.5, 46.2, 48.1, 50.0, 51.9, 53.8, and 55.7 degrees Celsius.
However, we hit quite a snag in our research when our PCR reactions inexplicably stopped working; we were using the same primers, samples, reagents, and PCR profile as a previously successful experiment. The last few weeks of summer and the first of the semester Dr. Brokaw and I had to backtrack in order to identify what element was hindering our progress. We systematically ran PCRs substituting out various reagents and ingredients of our experiments. After a number of consistent failures, we remixed our primers and revisited a procedure that had worked previously, to our relief we found bands in the gel once more (in the right base pair region to boot). We still aren’t sure, but it is possible that poor water quality was the problem.
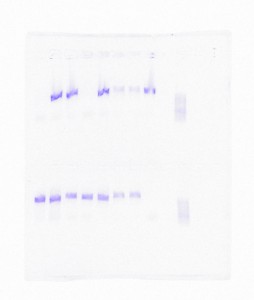
Gel of 15 DNA samples amplified with the remixed p484-p485 primers.
After moving past the hiccup in our research, I used the newly mixed primers (with good water) and ran another PCR with a gradient to try and get all the samples to show adequate amplification. With good results on the PCR, we preformed a cleaning using a Cycle Pure kit. Tanya and I quantified the DNA from our cleaned samples using a NanoDrop to make sure we retained our DNA and had adequate amounts. We will be sending off the samples to be sequenced later this week. When they return, our next step will be to start the DNA editing for the phylogenetic study.
We are also in the process of a DNA extraction from liver tissue to prepare new samples provided by Dr. Lee and his research assistants. The data should be quite interesting to compare with our previous samples as they were collected on opposite slopes of the Andes mountains.
