The month of August tested my ability to not only accomplish my own work, but to perform the tasks of my two research partners while they were away on vacation. The key words for the summation of my success were organization and documentation.
I began the month where I left off in July, with the attempt to perform a gel DNA recovery on tapY1. All I can say is frustration. Every time I performed this process with the appropriate kit, I received poor results on the Nanodrop (I was hoping to receive a nucleic acid concentration of at least 200ng/ul, but the highest I obtained was 50.0ng/ul). The gene sequence, recA, was also ready for gel DNA recovery, and to my dismay, the results displayed extremely low concentrations. Both tapY1 and recA were sent back to the stage of large-scale PCR using AccuTaq. The gene sequence, clpAS, finally reached the stage of moving from PCRs with Taq to PCRs with AccuTaq. First, I ran an AccuTaq PCR gradient to discover the proper annealing temperature. Second, I ran a large-scale AccuTaq PCR using the appropriate annealing temperature to amplify the 4.6kb product. Once the large-scale PCRs for tapY1, recA, and clpAS were all run, gel DNA recovery was attempted one final time.
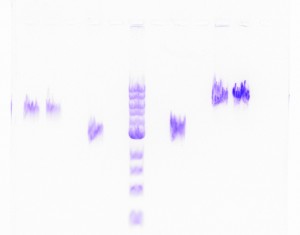
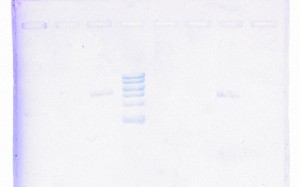
Yikes! Not only were the results extremely poor, but also, the gel run with the DNA recovered products produced fuzzy bands, all of which were the wrong size. Just when I started thinking progresses was being made, I learned that backtracking was the new trend of my research. I set up Nesting PCRs for tapY1, recA, and clpAS using their innermost primers and the gel recovered DNA as the template DNA. This protocol was used to confirm whether or not the recovered DNA came from the appropriate section of the genome. Thankfully, the correct band sizes were produced! I proceeded to large-scale PCR for tapY1, recA, and clpAS using the outermost primers and the gel recovered DNA. The gene sequence, recA, was successfully amplified, but unfortunately, tapY1 and clpAS were not. The recA product was cleaned-up with a PCR Inhibitor Removal Kit and stored in the freezer until ready to send-off for sequencing. Finally, I advanced tapY1 and clpAS to the stage where their products were able to be cleaned-up with the PCR Inhibitor Removal Kit.
When the gel was run with all the PCR purified products, tapY1 and recA both displayed multiple bands. Also, the bands were not tight, but very wavy. A serious decision was made to transition from using SYBR green gels to using ethidium bromide gels. Although the health risks are higher from ethidium bromide, the gel bands are much tighter, without drags. Finally, the day arrived when all of the DNA samples were ready to be sent off for sequencing. I diluted the PCR purified products for tapY1, recA, and clpAS, and I diluted all of the primers for each of the three genome regions. Sufficient organization, color-coding, and packing were insured to make the parts to the puzzle easy for the sequencing company to distinguish. How exciting it was to reach this initial goal of research! When the results returned, the company concluded that almost all of the data was bad. Future research will consist of comparing the returned sequences with predicted Geneious results to determine whether the sequences are usable or not.
A side project I worked on this month was constructing a replica-plating device out of a plastic cup, ring clamp, foam board, and a Kleenex. My design was a success, but the Kleenex hypothesis was not a sufficient replacement for the original velvet. The texture of the Kleenex was to thin and absorbed the media liquid, effecting the bacteria’s transfer onto a new TSA plate. Test will be run in the future to determine whether velvet will work on my device and allow adequate replica-plating. A huge discovery this month was that the deionized water in the microbiology lab and the research lab registered a pH of 1.9, instead of 7. Oh, no! No wonder so many test resulted in poor results! The acidity was killing the microorganisms. At least the mystery has been solved, and research can continue with a new sense of awareness of how important the pH of water really is! Summer research has been an exciting and rewarding experience. Now that school has begun, a new challenge I face is scheduling time to continue my research. I am excited to see how my research will progress during the month of September.