This has been quite a year for me and Samantha, and our research with Dr. Brokaw. We have learned a lot, about our research area in Oklahoma, about the ACU Biology Department (that has so graciously welcomed a couple crazy Ag majors tag along on their conference trips, Biology club meetings, and so on), and about how the research process works. It’s been fun! And this is the last blog I believe I’ll be posting, which is bittersweet. I’m thankful to Pursuit, to ACU, to Dr. Brokaw, and most of all, my awesome partner, Samantha, for this entire process.
So far, our results have been as expected: disturbance-loving plants took over the spill sites, and the less competitive species have been in areas of lower hydrocarbon concentrations and on our control sites. In my opinion, we reached our goals as best as we could – seeing as how our goals were mostly to get through the samples at least one time this year (there were 40+ samples that we were trying to get through, on top of being full time students with jobs and families).
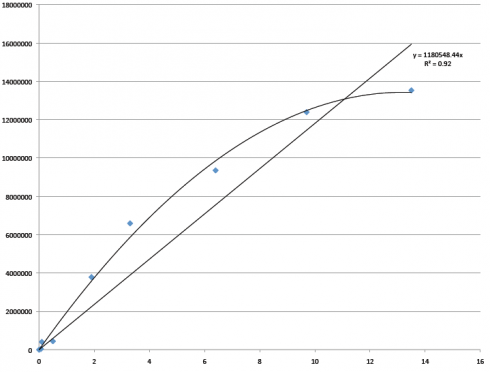
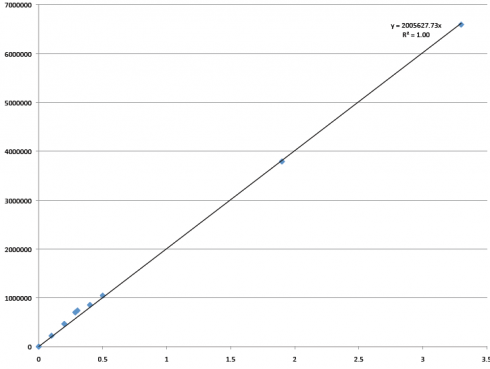
We smoothed out so many bumps in the road that will make way for the future: figuring out the fussy gas chromatograph, thinking we lost all of the research when I lost the thumb drive (it was mostly backed up, thankfully) and how to prevent that from happening again, coming up with a rhythm, and making decisions and alterations on so many little things that I can’t even remember them all now. Having these experiences, both traumatic and exciting, and getting through them successfully has encouraged me and enhanced my problem solving abilities. The extra activities I’ve gotten to participate in, like going to Hawley Middle School and talking to the kids for two years now, has helped me understand the importance of my research and how valuable it is to pass on the information and the interest of bioremediation. This alone has been an unexpected and pleasant surprise.
I am excited for the future of this project. I think a research endeavor like this, one that is about remediating an area that has been contaminated by a man-made accident, is really important for ACU and a great opportunity for its students. I will continue to work on this project for the next month or so, and hopefully in the fall I will be able to help with training another student to take over, since Samantha and I have graduated. I hope that the student is able to devote a lot of time to this, perhaps by securing their own grant, and are therefore able to work less (if at all) somewhere else; this is the only suggestion for change that I have – just having students that are available to spend a lot of time on the project. It’s the best way to keep consistency within the methodology of the project, and when it has worked best for us.
Bioremediation of a Tall Grass Prairie has been a great scholarly journey for me. Samantha and I have presented research at the ACU Undergraduate Research Festival and the Texas Academy of Science. Both were great opportunities and I learned a lot about what I do well and what I don’t. I hope that this research project and what it taught me will be a solid foundation for the research I hope to get to do in the future.